A collaboration between pure and applied mathematicians, pathologists and clinicians, has shown how sophisticated techniques from computational algebraic topology can be applied to understand the spatial distributions of immune cell subtypes in the tumour microenvironment.
The mathematical tool they showcase is multi-parameter persistent homology (MPH) landscapes, developed by a recent Centre for Topological Data Analysis student, Oliver Vipond. MPH landscapes is a stable and computable invariant for studying data with multiple parameters using topological data analysis (see a previous article on MPH landscapes by Urike Tillmann). This is the first study to apply MPH landscapes to genuine real world applications.
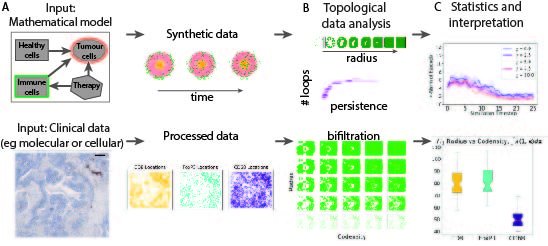
Using a mechanistic model, the team generate synthetic spatio-temporal data of immune cells infiltrating a tumour spheroid, where the ground truth is known (Figure 1A, top). They then apply MPH landscapes to the data and show how it surpasses single parameter persistent homology (Figure 1B). This synthetic study also validates the interpretation of MPH landscapes (Figure 1C).
Next, the team showcase MPH landscapes on slices of cancer tissue (Figure 1A, bottom). They show how MPH landscapes can quantify, characterise, and compare the distributions of different immune cell subtypes as they are infiltrating into tumours, and how these patterns change in different environmental conditions such as hypoxia (i.e., low oxygen levels). The team propose that one of the parameters, i.e., co-density in the bi-filtration (Figure 1B, bottom), is a good proxy for the oxygen level within the tissue. As the point clouds are too large for a single computation, they take multiple subsamples. Statistical analysis of the MPH landscapes suggests that infiltrating regulatory T cells have more prominent voids in their spatial patterns than macrophages (Figure 1C, bottom). This study highlights how Topological Data Analysis can integrate and interrogate data of different types and scales, e.g., immune cell locations and regions with differing levels of oxygenation.
This work is truly interdisciplinary: the mechanistic models generate synthetic data that is used as a ground truth to validate and interpret the utility of the methods, which gives greater confidence to apply them to study the immune/tumour microenvironment. This work highlights the power of MPH landscapes for quantifying, characterising, and comparing features within the tumour microenvironment in synthetic and real datasets and the benefits of working across disciplines.
You can read more about this work which appears in Proceedings of the National Academy of Sciences (PNAS), co-authored by Oxford mathematicians, clinicians and pathologists: Oliver Vipond, Joshua Bull, Philip Macklin, Ulrike Tillmann, Christopher Pugh, Helen Byrne and Heather Harrington (pictured top).