Oxford Mathematician Benjamin Walker talks about his work on the automatic identification of flagella from images, opening up a world of data-driven analysis.
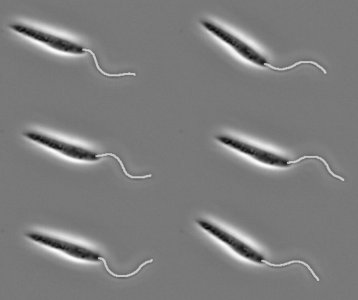
"Across the University of Oxford alone there are millions of images of microorganisms propelling themselves along with flagella, slender tail-like structures like those found on the sperm cells of most mammals. Studying the detailed motion of flagella is of intense scientific interest, not least amongst researchers in the fields of fertility medicine and human pathogens. Previously, flagella in each of these countless images would need to be traced (often by hand on a graphics tablet) by an unfortunate researcher before they can analyse the detailed motion of the fagellum. With even the more-sophisticated 'point-and-click' computerised methods, in general this still requires vast amounts of time and laborious effort. With artificial intelligence capable of accurately detecting cats in videos on YouTube, and a lot more besides, it seems that automation of this somewhat-simple task might be easily accomplished using the powerful computing resources available today.
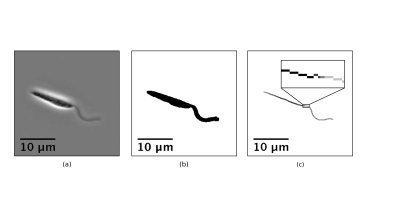
In fact, we needn't turn to complex algorithms or simulated intelligence at all to automate our identification and remove the need for a patient researcher. Instead we look at something fundamental in the biology of the flagellum, persistent across a wide range of organisms. Eukaryotic flagella, like those of mammalian sperm and the human parasites Trypanosomatidae, are all made up of the same structural backbone, referred to as the 9+2 axoneme, which gives them a well-defined and consistent width when viewed through typical microscopes. If we compare this to the rest of the cell, for example for the Trypanosome shown below in (a), the flagellum stands out as by far the largest portion of the microorganism that is a consistent thickness.
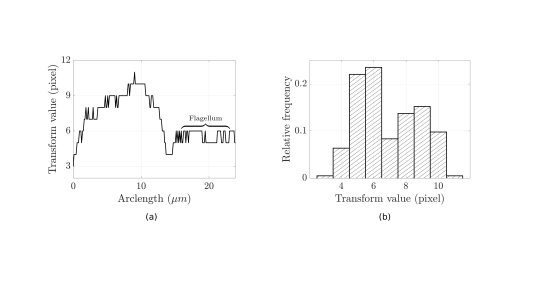
The width of the organism is something that can be easily quantified via a medial axis transform. The transform takes a binary representation of the whole cell (b), obtainable automatically, and reduces it down to a skeleton along the approximate midline of the cell (c). We then colour each point on this skeleton by the distance to the edge of the cell, with the wider parts corresponding to darker sections of the skeleton. Tracing out the skeleton from left to right, we can record the width of the cell at each point, as shown on the graph to the left below.
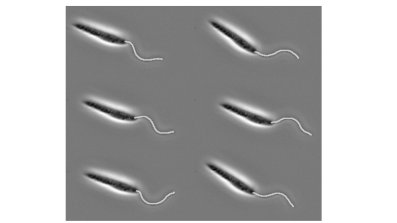
The flagellum can be easily identified by eye as the large region of approximately constant width. The histogram to the right in fact shows something more, that the flagellar width (approximately 6 pixels here) corresponds to the mode of all the cell widths. This observation leads to a remarkably simple scheme for automatically identifying flagella: take a medial axis transform, and keep only the regions corresponding to the modal width. We have tested this simple method on a sample of 150,000 images of a Trypanosome, orders of magnitude more data than previously analysed for this organism, and accurately captured the flagella of over 120 different cells, enabling population-level studies of flagellar movement.
Thus, with only a simple image transform and exploiting a conserved structural feature of flagella, this automated methodology could enable rapid quantification of large image datasets, freeing up researchers to analyse data rather than trace it out."