Topological data analysis enables deep quantification of vascular responses to drug and radiation treatment in 3D tumours. Oxford Mathematician Bernadette Stolz explains.
Advances in imaging techniques enable high resolution 3D visualisation of vascular networks over time and reveal abnormal structural features such as twists and loops. The quantification of the 3D architecture is important because vessel structure affects vessel function (i.e., delivery of oxygen, nutrients, and therapies).
Existing analyses have quantified structural features, including vessel density, number of vessels and branching points, and highlighted their relevance for predicting disease progression and response to treatment. Such spatially averaged summaries focus on a single spatial scale and lose information from detailed 3D renderings. Moreover, they do not account for vessel connectivity, or higher order topological features such as loops and voids; the latter correspond to avascular tumour regions associated with reduced patient survival and poor responses to therapy. Therefore, more detailed, automated and reproducible methods for quantifying vessel networks are needed, which may provide future benefit to basic research, clinical assessments and treatment planning.
Topological data analysis (TDA), the mathematical field that studies ‘shape’ of data, has expanded from theory to applications through advances in computation and integration with machine learning. As myself and my colleagues Heather Harrington and Helen Byrne highlight in our recent paper Multiscale Topology Characterises Dynamic Tumour Vascular Networks [SKMMLMBH], TDA can characterise the geometric, spatial and temporal organisation of vascular networks. We propose two topological lenses to study vasculature, which capture inherent multi-scale organisation and vessel connectivity invisible to existing methods, and which surpass the single scale analysis of existing methods. The inputs to TDA are the full 3D structures of spatial networks obtained from intravital and ultramicroscopy imaging modalities, and outputs are interpretable multiscale topological descriptors, such as tortuosity, spatio-temporal variation in the number of loops and size of avascular regions (voids), which are inaccessible from analysis of 2D representations (eg slices or image projections). The two topological lenses, or filtrations, that we propose are the ‘radial filtration’ and ‘α -complex filtration’ (see figure below).
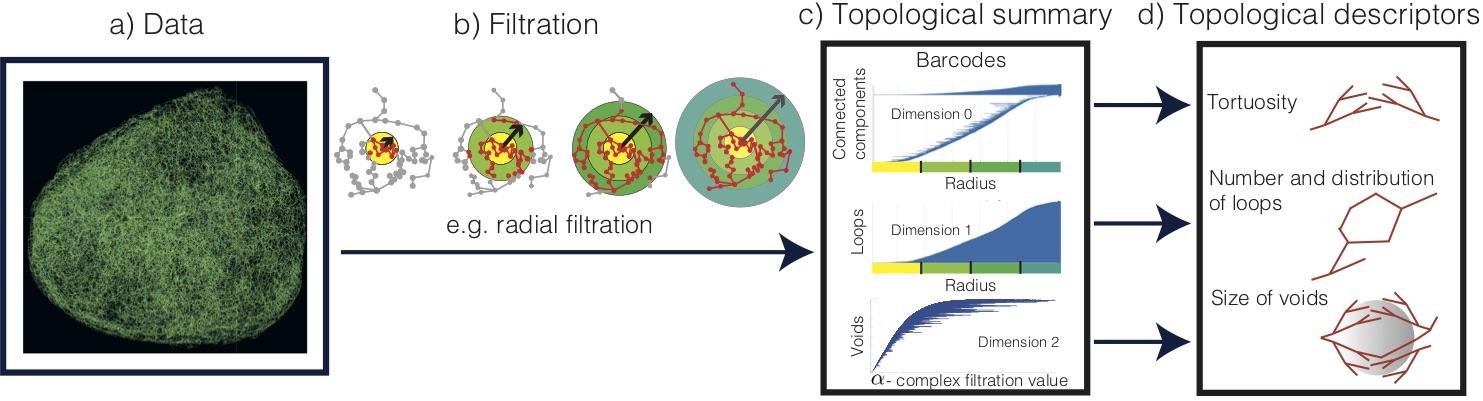
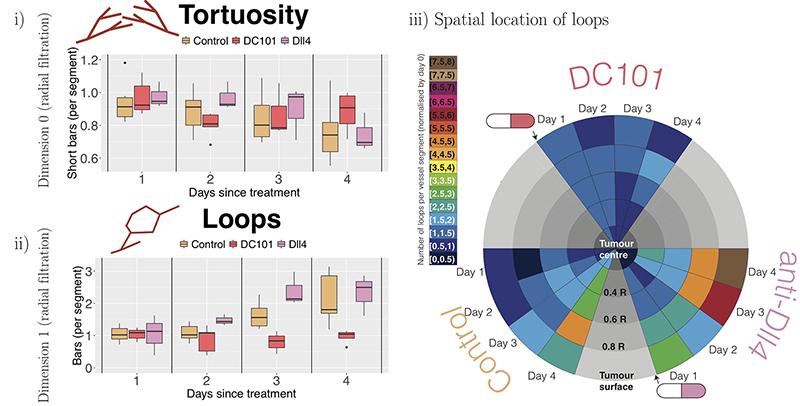
This unprecedented quantification enables us to validate known qualitative trends in network responses, such as transient vessel normalization (i.e., dynamic changes in tortuosity and loops), in response to antibodies that modulate vessel sprouting. The topological descriptors computed from intravital data of tumour blood vessels, shown in the figure below, confirm the known effects of the vascular targeting agents DC1010 (reduces vascular sprouting) and anti-DII4 (increases vascular sprouting). Furthermore, TDA quantifies the unknown effects of radiotherapy on vessel architecture.
References:
[SKMMLMB] Multiscale Topology Characterises Dynamic Tumour Vascular Networks. Stolz BJ, Kaeppler J, Markelc B, Braun F, Lipsmeier F, Muschel RJ, Byrne HM, Harrington HA.


