Globally kidney disease is forecast to be the 5th leading cause of death by 2040, and in the UK more than 3 million people are living with the most severe stages of chronic kidney disease. Chronic kidney disease is often due to autoimmune damage to the filtration units of the kidney, known as the glomeruli, which can occur in lupus, a disease which disproportionally affects women and people of non-white ethnicities, groups often underrepresented in research. Treatment options are limited, can have life threatening side-effects and often don’t slow the disease, which can then progress to end stage, requiring dialysis or a kidney transplant.
There is an urgent need to develop safer, more effective and personalised treatments for these kidney disorders, and in Oxford, researchers are using new technologies to address this problem. Tools that allow scientists to understand gene expression in individual cells, after separating them out from a kidney sample, have been around for about 10 years, but very recently new techniques are starting to reveal these cell signatures in the tissue context. This means the cells can be considered as part of their local neighbourhood, showing how cells move and signal to each other. This approach is exciting for autoimmune kidney diseases like lupus, because the kidney is complex with more than 30 types of cell, and in lupus these are joined by multiple immune cell types. Yet how these interactions lead to kidney damage is not well understood.
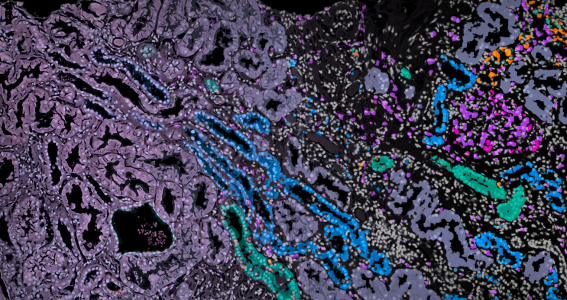
Aneesha Bhandari and Katherine Bull in the Nuffield Department of Medicine in Oxford have been using these new spatial methods to look at lupus kidney disease, but Aneesha and Katherine encountered a problem; the spatial information is at unprecedented subcellular resolution, but very sparse and noisy, so deciding what type of cells they had found, and where the boundary of each cell lies, was challenging. They knew there must be a way to dynamically use the local expression information to improve this, but how?
Luckily a conversation with Oxford Mathematicians Heather Harrington (also Max Planck Institute of Molecular Cell Biology and Genetics), Ulrike Tillmann and Katherine Benjamin, revealed an elegant solution. Their research in topological data analysis identifies spatial patterns in data across different parameters. Together the teams developed a new method, TopACT, to apply this mathematics to the spatial kidney data. They were able to reveal hidden patterns in the lupus kidney, with immune cells circling the glomerular regions. The approach turns out to work on a range of spatial platforms, important as these technologies are moving fast, and could be applied in the future to 3-dimensional data.
"This is a really interdisciplinary effort between mathematics and biology, allowing us to see granular detail and hidden patterns of inflammation in the kidney in lupus. These tools are an important step towards developing more targeted ways to treat this complex disease" - Katherine Bull
"We are excited that this new spatial transcriptomics technology required us to develop novel mathematical approaches, building upon state of the art topological data analysis. In this work we identified spatial locations of sparse cells and then generated a hypothesis of a ring of immune cells in mouse kidney tissue, which was experimentally verified. Trying to understand this complex biological data offers interesting and challenging mathematical opportunities.” - Heather Harrington
You can read more about the work, published in Nature today.
Image: mathematical methods reveal complex cell patterns in high-resolution kidney data