Research: During an outbreak
When an infectious disease outbreak is ongoing, our research group is interested in how mathematical models can be used to guide control interventions.
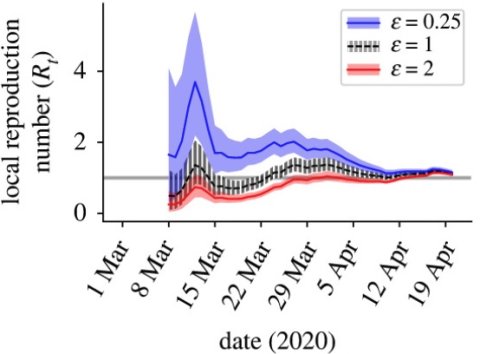 We have worked on methods for estimating the "R number" - the expected number of infections generated by each infected individual. This quantity can be tracked during outbreaks to monitor ongoing transmission and the effectiveness of interventions. However, many common features of outbreak data can make estimation of the R number more challenging. For example, outbreak data might be aggregated temporally, with weekly numbers of cases being recorded rather than daily numbers of cases, and key features of infections may not be considered (e.g., whether individuals were infected within or outside the local population). We have developed methods to address these issues, enabling more robust estimates of the R number to be obtained.
We have worked on methods for estimating the "R number" - the expected number of infections generated by each infected individual. This quantity can be tracked during outbreaks to monitor ongoing transmission and the effectiveness of interventions. However, many common features of outbreak data can make estimation of the R number more challenging. For example, outbreak data might be aggregated temporally, with weekly numbers of cases being recorded rather than daily numbers of cases, and key features of infections may not be considered (e.g., whether individuals were infected within or outside the local population). We have developed methods to address these issues, enabling more robust estimates of the R number to be obtained.
In recent work funded by the World Health Organization, we have been investigating the optimal COVID-19 booster vaccination strategy to implement in different countries. While such analyses have been undertaken for high income countries such as the UK, less attention has been directed towards low and middle income countries. Since booster vaccine supply is limited in every country, it is essential to consider who to prioritise for booster vaccination. Our latest modelling results suggest that prioritising older (and other vulnerable) individuals for booster vaccination leads to the best possible public health outcomes in every country considered, aligning with findings earlier in the pandemic for initial COVID-19 vaccinations.
Publications from our group relating to using models to understand transmission and guide control interventions when outbreaks are underway include:
- Ogi-Gittins I, Hart WS, Song J, Nash RK, Polonsky J, Cori A, Hill EM, Thompson RN. A simulation-based approach for estimating the time-dependent reproduction number from temporally aggregated disease incidence time series data (2024) Epidemics, 47:100773. (Available here)
- Doyle NJ, Cumming F, Thompson RN, Tildesley MJ. When should lockdown be implemented? Devising cost-effective strategies for managing epidemics amid vaccine uncertainty (2024) PLoS Comp. Biol. 20:e1012010. (Available here)
- Ogi-Gittins I, Steyn N, Polonsky J, Hart WS, Keita M, Ahuka-Mundeke S, Hill EM, Thompson RN. Simulation-based inference of the time-dependent reproduction number from temporally aggregated and under-reported disease incidence time series data (2025) Phil. Trans. Roy. Soc. A, 383:20240412. (Available here)
- Hart WS, Miller E, Andrews NJ, Waight P, Maini PK, Funk S, Thompson RN. Generation time of the alpha and delta SARS-CoV-2 variants: an epidemiological analysis (2022) Lancet Inf. Dis. 22:603-610. (Available here)
- Hart WS, Abbott S, Endo A, Hellewell J, Miller E, Andrews N, Maini PK, Funk S, Thompson RN. Inference of the SARS-CoV-2 generation time using UK household data (2022) eLife 11:e70767. (Available here)
- Creswell R, Augustin D, Bouros I, Farm HJ, Miao S, Ahern A, Robinson M, Lemeneul-Diot A, Gavaghan DJ, Lambert BC, Thompson RN. Heterogeneity in the onwards transmission risk between local and imported cases affects practical estimates of the time-dependent reproduction number (2022) Phil. Trans. Roy. Soc. A, 380:20210308. (Available here)
- Hart WS, Maini PK, Thompson RN. High infectiousness immediately before COVID-19 symptom onset highlights the importance of continued contact tracing (2021) eLife, 10:e65534. (Available here)
- Thompson, RN. Epidemiological models are important tools for guiding COVID-19 interventions (2020) BMC Med., 18:152 (Available here)
- Hart WS, Hochfilzer LFR, Cunniffe NJ, Lee H, Nishiura H, Thompson RN. Accurate forecasts of the effectiveness of interventions against Ebola may require models that account for variations in symptoms during infection (2019) Epidemics, 29:100371 (Available here)
- Thompson RN, Stockwin JE, van Gaalen RD, Polonsky JA, Kamvar ZN, Demarsh PA, Dahlqwist E, Li S, Miguel E, Jombart T, Lessler J, Cauchemez S, Cori A. Improved inference of time-varying reproduction numbers during infectious disease outbreaks (2019) Epidemics, 29:100356 (Available here)
Return to our Research page by clicking here.

