Investigating Graph-Based Machine Learning Methods in Drug-Discovery
- Researcher: Markus Dablander
- Academic Supervisors: Renaud Lambiotte, Garrett Morris
- Industrial Supervisor: Thierry Hanser
Background
Machine learning techniques based on graph-shaped input data have recently shown great promise for the prediction of molecular properties and for the in silico discovery of novel molecules with desired qualities. Lhasa are interested in exploring how these techniques could be used to enhance the streamlining and standardisation of drug development processes, the reduction of animal testing, and the computational prediction of toxicity, metabolic fate and chemical degradation of molecular compounds. In our project, we explore the potential of both classical and neural machine learning methods operating on molecular graph structures for the acceleration of chemical research and the enhancement of computer-aided drug discovery.
We put a particular focus on the creation of novel techniques for the automatic transformation of molecular graphs into informative vectors of real numbers which can be fed into machine learning pipelines for the purpose of molecular property prediction. The process of representing molecules via carefully desgined feature vectors which contain relevant information to help predict a given end-point (such as toxicity) is known as molecular featurisation (see Figure 1) and has traditionally been a bottleneck for the performance of predictive algorithms in computational chemistry. We are interested in addressing this obstacle by developing and testing novel techniques for the automatic extraction of meaningful features from molecular graphs.
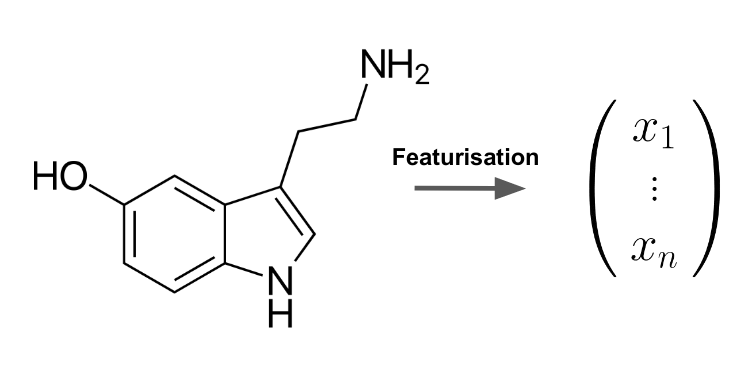
Figure 1: Molecular featurisation is a key step in computational chemistry and describes the process of turning a molecular structure into an informative vector of real numbers.
Besides this, we also focus on other bottlenecks of predictive models in molecular machine learning such as activity cliffs. Activity cliffs are formed by pairs of molecules with highly similar structures but large differences in activity against a given pharmacological target of interest. The presence of activity cliffs in molecular data sets forms a great challenges for common machine-learning algorithms and can severely limit their predictive abilities. At the same time, knowledge about activity cliffs can have important implications for compound optimisation and the elucidation of structure-activity relationships. We are working on novel methods which can decrease the negative effects of activity cliffs on predictive models and which can capitalise on the chemical knowledge encapsulated in them. As part of this, we are developing new graph-based methods for the prediction of activity cliffs in chemical space.
In our research process we heavily (but not exclusively) experiment with techniques from deep learning and artificial intelligence such as graph neural networks, self-supervised learning and transfer learning. Our overarching goal is the improvement of algorithms from molecular machine learning via the construction of novel molecular representations and the identification and amelioration of performance bottlenecks.
Outcomes
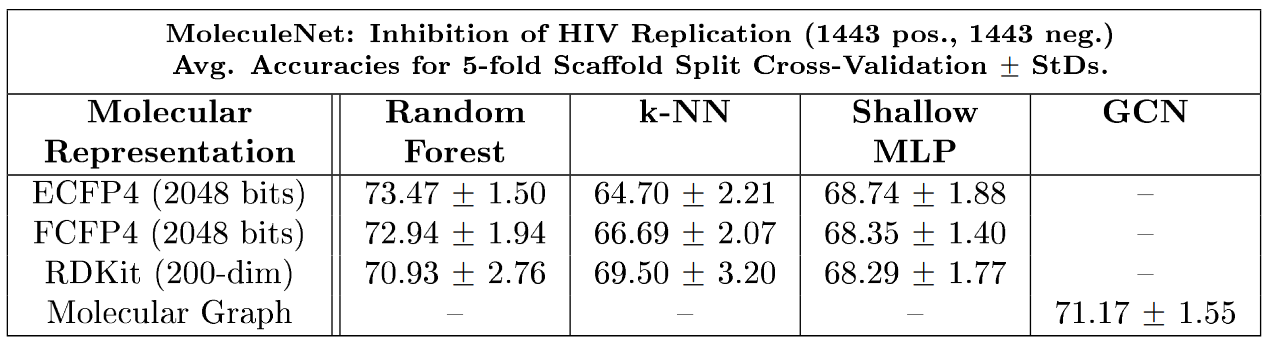
In one of our first computational experiments, we tested how the performance of molecular features which are automatically extracted via a graph convolutional network (GCN) compare to classical features based on circular fingerprint techniques (ECFPs and FCFPs) which encode the absence or presence of molecular substructures in high-dimensional binary feature vectors. When compared to fixed circular fingerprints, the more modern GCNs open up a variety of new feature-learning possibilities such as task-adaptivity, transfer learning and domain adaptation. Next to ECFPs/FCFPs and GCNs, we also tested the performance of molecular featurisations based on global molecular descriptors computed via the well-known RDKit Python chemoinformatics package.
Figure 2: Results of a computational study to compare the predictive performance of classical and modern molecular featurisation techniques for the identification of small molecules with inhibitory effect on HIV replication.
We considered a balanced binary molecular classification problem consisting of molecular compounds which are either active or inactive as inhibitors of HIV replication. In spite of various claims in the recent literature that featurisation methods based on graph neural networks are generally superior to circular fingerprints, we observed in our results (see Figure 2) that the much simpler and computationally cheaper circular fingerprint features lead to a performance comparable to the one obtained via graph neural networks. These observations support our hypothesis that the feature-extraction capabilities of graph neural networks are in fact not (yet) consistently superior to featurisation techniques based on classical, simple-to-compute molecular descriptors such as circular fingerprints. The results thus suggest the necessity for breakthroughs in the field of molecular featurisation in order to further increase model performance.
Future Work
One of our next steps will be to study the intersection of molecular featurisation and activity cliff prediction. Current molecular featurisations are largely based on a principle of structural similarity, whereby structurally similar molecules end up having similar feature vectors. While generally reasonable, this principle can impair the performance of machine learning algorithms when trying to predict the activities of structurally similar molecules with highly different activity levels. To overcome this obstacle, we intend to experiment with a novel type of graph neural network architecture for the creation of activity-cliff-sensitive molecular feature vectors. Combining this novel graph-based neural architecture with the idea of activity-cliff aware feature learning has the potential to lead to new and more powerful molecular features.